Abstract
Simple repetitive DNA sequences are a widespread and abundant feature of genomic DNA. The following features characterize such sequences: (1) they typically consist of a variety of 1-10 base repeat motifs, but may also include much larger repeats; (2) larger repeating units often include shorter ones within them; (3) long polypyrimidine and poly-CA tracts are often found; and (4) tandem arrangements of closely related motifs are often found. We propose that slipped strand mismatch events, together with unequal crossing over, can easily explain all of these features.
The frequent occurrence of long tandem repeats of particular motifs (polypyrimidine and poly-CA tracts) appears to be the result of non-random patterns of nucleotide substitution. We argue that the intrahelical process of slipped strand mismatching is much more likely to be the major factor in the initial expansion of short repeat motifs and that, after the initial expansion, simple tandem repeats may be predisposed to further expansion by unequal crossover or other interhelical events due to their propensity for mismatching.
Evidence is presented that single base repeats (the shortest possible motifs) are represented by longer series in mammalian introns than would be expected randomly, supporting the idea that SSM may be a ubiquitous force in the evolution of the eukaryotic genome. Thus, simple repetitive sequences may represent a natural ground state of DNA not selected for coding functions.
Key points
- Altered gene expression is the result of SSM and, depending on where the increase or decrease in short repeat sequences occurs relative to the promoter, will be regulated at the level of transcription or translation. The result is an ON or OFF phase of a gene or genes.
- SSM can result in an increase or decrease in the number of short repeat sequences. Short repeat sequences are from 1 to 7 nucleotides and can be homogeneous or heterogeneous repetitive DNA sequences.
- Transcriptional regulation can occur if the repeats are located in the promoter region at the RNA polymerase binding site, -10 and -35 upstream of the gene(s).
- SSM induces transcriptional regulation by changing short repeat sequences located outside the promoter. If there is a change in the short repeat sequence, it can affect the binding of a regulatory protein, such as an activator or a repressor.
Key terms
Slipped strand mismatch: a process that produces a mismatch of short repeat sequences between the parent strand and the daughter strand during DNA synthesis.

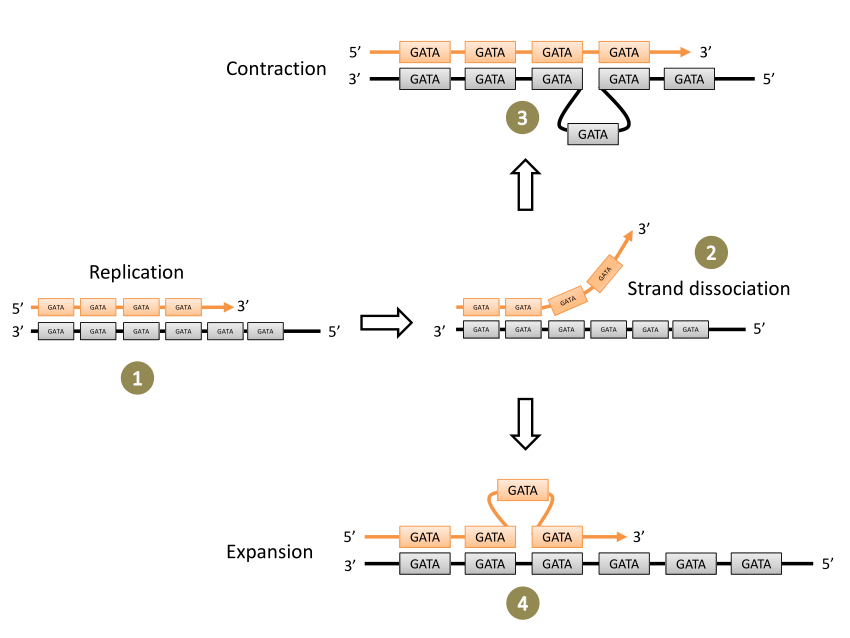
Slipped strand mismatch (SSM) is a process that results in the mispairing of short repeated sequences between the mother and daughter strand during DNA synthesis. This RecA-independent mechanism can occur during DNA replication or DNA repair and can be on the leading or lagging strand and can result in an increase or decrease in the number of short repeat sequences. Short repeat sequences are from 1 to 7 nucleotides and can be homogeneous or heterogeneous repetitive DNA sequences.
Altered gene expression is the result of SSM and, depending on where the increase or decrease in short repeat sequences occurs relative to the promoter, will be regulated at the level of transcription or translation. The result is an ON or OFF phase of a gene or genes. Transcriptional regulation occurs in several ways. One possibility is if the repeats are located in the promoter region at the RNA polymerase binding site, -10 and -35, upstream of the gene(s). The opportunistic pathogen H. influenza has two divergently oriented promoters in the gene shift and hifB fimbriae.
Overlapping promoter regions have TA dinucleotide repeats at -10 and -35 sequences. Via SSM, the TA repeat region can undergo the addition or subtraction of TA dinucleotides, resulting in the reversible ON or OFF phase of hifA and hifB transcription. The second way that SSM induces transcriptional regulation is by changing short repeat sequences located outside of the promoter. If there is a change in the short repeat sequence, it can affect the binding of a regulatory protein, such as an activator or a repressor. It can also give rise to differences in the post-transcriptional stability of the mRNA.